seqscreen
functional screening of pathogenic sequences
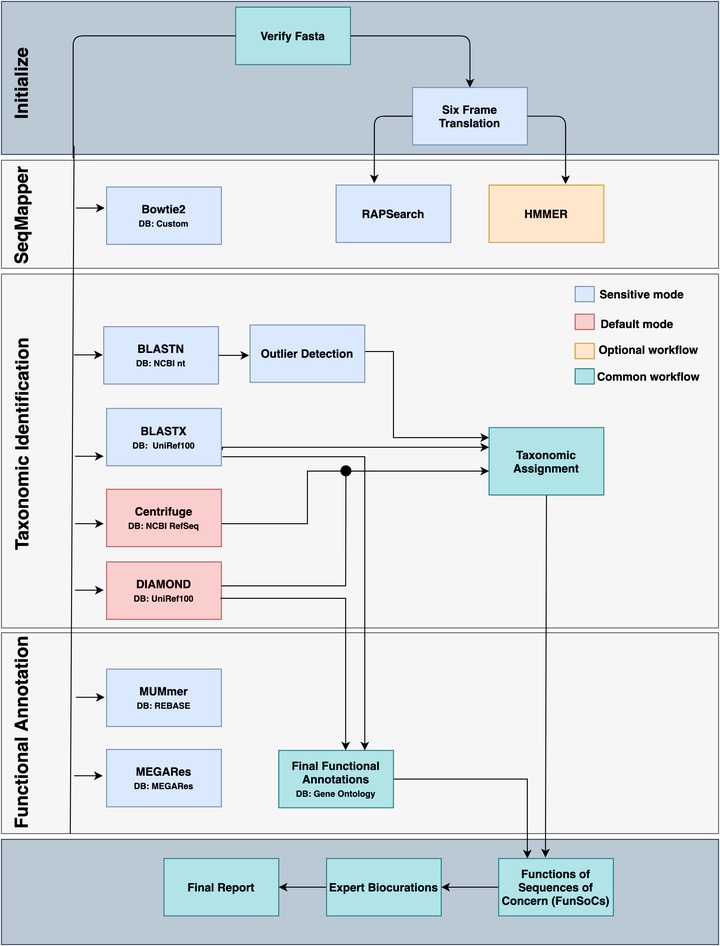
SeqScreen is a tool for characterizing the pathogenicity of short nucleotide sequences. It first gathers taxonomic and functional information on the input sequence and then utilizes this information to label the input necleotide sequence with Functions of Sequences of Concerns (FunSocs). FunSocs serve as abstractions of the underlying threat processes and as indicators of the threat level of a certain sequence. Examples of FunSocs include “disable organ” and “Cytotoxicity”.
As SeqScreen uses taxonomic and functional information to predict a necleotodie sequence’s pathogenicity, it is important to verify that SeqScreen is able to correctly identify the input sequence’s taxonomic and functional information. To this end, I curated a data set of short nucleotide sequences and benchmarked SeqScreen’s taxonomic and functional identification ability against software developed by other researchers. With the insights provided by my benchmarking pipeline, through trial and error, we were able to improve SeqScreen’s performance, making SeqScreen one of the most sensitive and accurate tools for taxonomic and functional indentification.
The ultimate goal of SeqScreen is to predict the pathogenicity of novel short nucleotide sequences. Previous attempts at this goal primarily relies on mapping a novel sequence to a known sequence in the database. I demonstrated the limitations of this approach by showing that if the novel input sequence is harmful and highly similar to a benign sequence in the database, the existing tools tend to map the input to its benign counterpart, thus failing to correctly characterzing the pathogenicity of the input sequence. On the other hand, I showed that SeqScreen is able to distinguish the novel malignant sequences, as they are labeled with FunSocs that are absent in their benign counterparts, thereby proving the usefulness and robustness of the new pathogenicity characterization paradigm introduced by SeqScreen.
Here is the link to an overview of SeqScreen on Treangen Lab’s webiste.